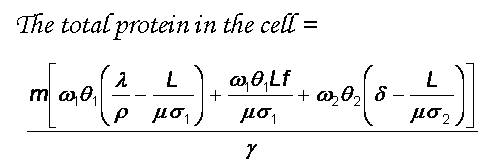The Lim lab combines theory and experiments to gain quantitative insight into the fundamental processes involved in gene regulation.
Our research is essential to understand the functional basis of genome and gene network evolution, to identify new strategies for treating bacterial infections, and to develop novel components that can be used in synthetic biology.
There are currently three major research areas.
1) Genome organization
Since the earliest days of genetics it was recognized that genes are located at non-random positions on the chromosome. For example, genes in the same pathway are often located next to one another on the chromosome. Our research examines the functional significance of this and other organizational features of genomes.
Fundamental relationship between operon organization and gene expression. Lim HN, Lee Y, Hussein R. Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10626-31.
2) Genetic switches
Bacteria have evolved several different classes of molecular switches that turn gene expression either fully on or fully off (i.e. like an ordinary light switch). Genes that are controlled in this manner, as opposed to genes that are expressed at varying levels (i.e. like a dimmer switch), are commonly involved in bacterial pathogenesis and surival during stress conditions. Intriguingly, these genetic switches often turn on and off randomly.
We are examining whether the different classes of molecular switches have different switching properties and the impact of random switching on bacterial survival and pathogenesis.
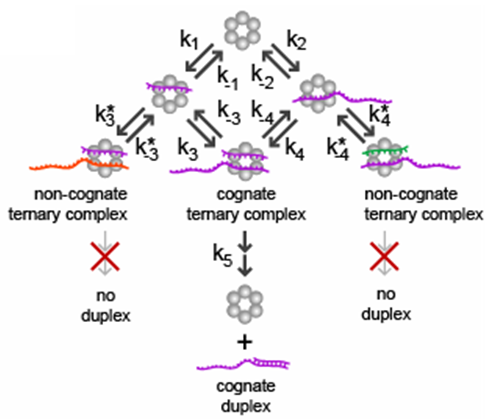
3) Small RNAs
Most organisms can regulate the expression of genes using small RNAs (sRNAs) and/or proteins acting as transcription factors. We are interested in why these two different types of signaling molecules have evolved - do sRNAs and proteins have unique properties?