Poster presented during the 79th Annual Meeting of
the American Society of Mammalogists,
University of Washington, Seattle, WA, 20-24 June
1999. Abstract # 89
Phylogenetic relationships among
echimyid rodents combining morphological and molecular data
Yuri L. R. Leite
Museum of Vertebrate Zoology, 3101 Valley Life Sciences
Building, University of California,
Berkeley CA 94720-3160. E-mail: yleite@socrates.berkeley.edu.
INTRODUCTION
Neotropical spiny rats, family Echimyidae (sensu
McKenna
& Bell 1997), are the most taxonomically, ecologically, and morphologically
diverse of all living hystricognath rodents. The earliest fossils date
from the Late Oligocene (25 MYBP), but modern taxa only appeared in the
fossil record in the Late Miocene (6.8-9 MYBP). Today, there are at least
70 species distributed from Central to South America, occupying a wide
variety of habitats and presenting a broad range of dietary and locomotory
habits. Despite this diversity, systematic studies of the group are still
in their infancy, and the evolutionary relationships among the genera are
still unclear.
Lara, Patton, & da Silva (1996) concluded that
a nearly simultaneous diversification of the most recent echimyids is likely
the explanation for the pattern of divergence found in a phylogenetic analyses
of the complete sequence of the mitochondrial gene cytochrome b (Figure
1). Their opinion was supported by the lack of resolution for relationships
among genera (polytomy); contrasted with strong support both above and
bellow this polytomy. Nevertheless, as pointed out by these authors, this
star-phylogeny hypothesis is potentially falsifiable by examining more
taxa and other markers.
My main goal is to test the star-phylogeny hypothesis
for the echimyid diversification. This can be achieved by adding molecular
data from different genes and by using morphological data, which allow
the inclusion of more taxa.

METHODS
Molecular data:
A total of 31 specimens were sequenced, including
27 echimyids belonging to12 different genera, plus 3 outgroup genera. The
same DNA extracts used by Lara, Patton, & da Silva (1996) were used
as templates for PCR amplification. The complete 12S gene (851 bp) was
amplified using three pairs of primers, and the last 581 bp of the 16 S
gene was amplified using two primers. Automated sequencing in both directions
was performed in an ABI Prism 377 DNA Sequencer, using the same sets of
PCR primers. Sequences were aligned by eye, based on the secondary structures
of the corresponding large and small subunits of the ribosomal RNA in the
mitochondria. Additional sequences from the genera Mus, Rattus,
Cavia,
Myocastor,
Capromys,
and Proechimys were downloaded from Genbank.
Morphological data:
Fourteen out of nineteen living echimyid genera
were included in the morphological analysis. The caviomorphs Coendou,
Myoprocta,
and Ctenomys were used as outgroups. The data matrix was assembled
using diagnostic traits previously described in the literature, characters
used in cladistic analysis of other rodent groups, and new features assessed
from direct examination of the specimens. The final data matrix consisted
of 41 characters, most of them related to tooth and skull morphology.
Phylogenetic analyses:
Parsimony analyses were performed using the computer
program PAUP* 4.0b2a (Swofford, 1997). Heuristic searches were conducted
using the tree-bisection-reconnection algorithm, via random stepwise addition
with 10 replicates. Character states were optimized using the accelerated
transformation option. The robustness of clades within trees was assessed
using bootstrap analysis (at least 100 replicates and 10 random sequence
additions per replicate). Pairwise sequence divergences were calculated
using the Kimura 2-parameter model.
RESULTS
Both 12S and 16S produced a polytomy for intra-generic
relationships among echimyids:
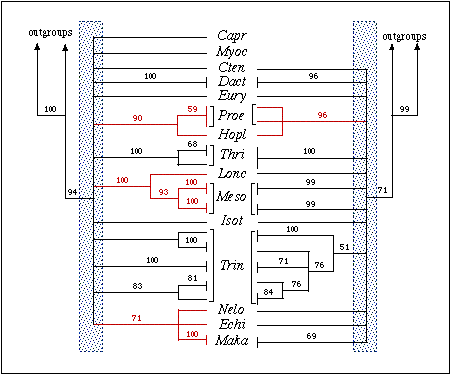 Figure 2.: Phylogenetic relationships among echimyid
genera based on sequences of the 12S (left) and 16S (right) genes. Both
are bootstrap consensus trees based on 1,000 replicates. Numbers are bootstrap
values above 50%. Outgroups taxa used were Mus, Rattus, Cavia,
Myoprocta,
and Coendou. Clades in red denote strongly supported sister-group
relationships between genera. The shaded area indicates the region of the
tree with no resolution (polytomy). Note that in the 12S tree the polytomy
includes the coypu Myocastor (considered an echimyid, especially
by paleontologists) as well as the hutia (Capromys) and the tuco-tuco
(Ctenomys), both non-echimyid octodontoids.
Figure 2.: Phylogenetic relationships among echimyid
genera based on sequences of the 12S (left) and 16S (right) genes. Both
are bootstrap consensus trees based on 1,000 replicates. Numbers are bootstrap
values above 50%. Outgroups taxa used were Mus, Rattus, Cavia,
Myoprocta,
and Coendou. Clades in red denote strongly supported sister-group
relationships between genera. The shaded area indicates the region of the
tree with no resolution (polytomy). Note that in the 12S tree the polytomy
includes the coypu Myocastor (considered an echimyid, especially
by paleontologists) as well as the hutia (Capromys) and the tuco-tuco
(Ctenomys), both non-echimyid octodontoids.
The star-phylogeny persists when sequence data
from 12S, 16S and cyt b are combined:
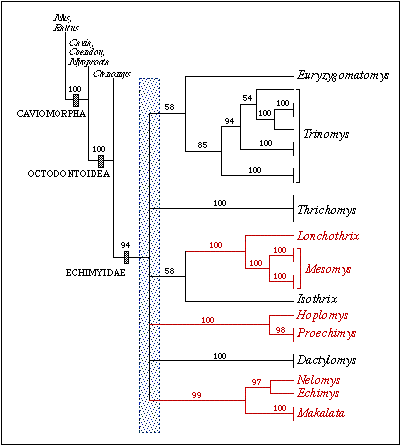 Figure 3: Phylogenetic relationships among
12 echimyid genera based on the combined sequences of the cyt b, 12S, and
16S genes(2,457 bp). This is a bootstrap consensus tree based on 1,000
replicates. Numbers are bootstrap values above 50%. The three clades in
red denote strongly supported sister-group relationships between genera.
In addition, there is very weak support for a Trinomys-Euryzygomatomys
clade and an Isothrix-(Lonchothrix-Mesomys) clade.
The shaded area indicates the region of the tree with no resolution (polytomy).
Figure 3: Phylogenetic relationships among
12 echimyid genera based on the combined sequences of the cyt b, 12S, and
16S genes(2,457 bp). This is a bootstrap consensus tree based on 1,000
replicates. Numbers are bootstrap values above 50%. The three clades in
red denote strongly supported sister-group relationships between genera.
In addition, there is very weak support for a Trinomys-Euryzygomatomys
clade and an Isothrix-(Lonchothrix-Mesomys) clade.
The shaded area indicates the region of the tree with no resolution (polytomy).
Morphological data offer some resolution, but
most relationships are still unresolved:
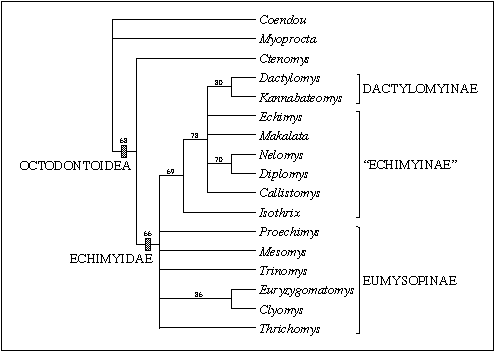 Figure 4: Phylogenetic relationships among 14 echimyid
genera based on 41 morphological characters. This is a bootstrap consensus
tree based on 1,000 replicates. Numbers are bootstrap values above 50%.
The current taxonomic classification, indicated at the right, suggests
the paraphyly of the subfamily "Echimyinae". The results support the distinction
between the recently described painted tree rat Callistomys (Emmons
& Vucetich 1998) and the brush tailed rat Isothrix, although
its relationship to other "echimyines" is still equivocal.
Figure 4: Phylogenetic relationships among 14 echimyid
genera based on 41 morphological characters. This is a bootstrap consensus
tree based on 1,000 replicates. Numbers are bootstrap values above 50%.
The current taxonomic classification, indicated at the right, suggests
the paraphyly of the subfamily "Echimyinae". The results support the distinction
between the recently described painted tree rat Callistomys (Emmons
& Vucetich 1998) and the brush tailed rat Isothrix, although
its relationship to other "echimyines" is still equivocal.
CONCLUSIONS
-
The results presented here corroborate the star-phylogeny
hypothesis. A protein-coding gene (cyt b), two ribosomal RNA genes (12S
and 16S), and morphological data were unable to resolve the polytomy, suggesting
that it reflects the true cladogenetic history of the group.
-
The molecular results unequivocally support three
clades within echimyids: (Lonchothrix-Mesomys), (Proechimys-Hoplomys),
and (Makalata(Nelomys-Echimys)). These results indicate
that the split between these lineages occurred after the basal split of
most living echimyid genera.
-
If we translate the Proechimys-Hoplomys clade
into a classification, then Trinomys must be recognized as a full
genus and not as a sub-genus of Proechimys, as suggested by Lara,
Patton & da Silva (1996).
-
The subfamily "Echimyinae" is paraphyletic according
to the morphological data. The tree rats are more closely related to the
bamboo rats (Dactylomyinae) than to the brushed tailed rat Isothrix.
FUTURE DIRECTIONS
The results show substantial support for the star-phylogeny
hypothesis, especially if we assume that mtDNA evolves in a roughly clock-like
manner within echimyids. However, if nucleotide substitutions occurred
at relatively low rates during the time when the clades containing the
extant genera diverged, then cladogenesis was not necessarily simultaneous.
To further address this question, additional sequences from independent
sources like the nuclear genome would be very important. Further, a cladistic
analysis based on morphological traits that includes fossil material may
potentially unveil these relationships, especially considering that the
family is relatively old and 30 out of 49 echimyid genera are extinct.
ACKNOWLEDGEMENTS
Financial support for this research was provided
by the Brazilian Research Council (CNPq), the Museum of Vertebrate Zoology,
and NSF grants to J. L. Patton. Thanks to Jim Patton, Eileen Lacey, Peg
Smith, Diogo Meyer and Leonora Pires Costa for helpful discussions and
continuous support.
REFERENCES
Emmons, L. H., & M. G. Vucetich. 1998. The
identity of Winge's Lasiuromys villosus and the description
of a new genus of echimyid rodent (Rodentia: Echimyidae). American Museum
Novitates, 3223:1-12.
Lara, M. C., J. L. Patton, & M. N. F. Da Silva.
1996. The simultaneous diversification of South American echimyid rodents
(Hystricognathi) based on complete cytochrome b sequences. Molecular Phylogenetics
and Evolution, 5:403-413.
McKenna, M. C. & S. K. Bell. 1997. Classification
of mammals above the species level. Columbia University Press, New York.
Swofford, D. L. 1999. PAUP*. Phylogenetic Analysis
Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland,
Massachusetts.
[Back]
|



