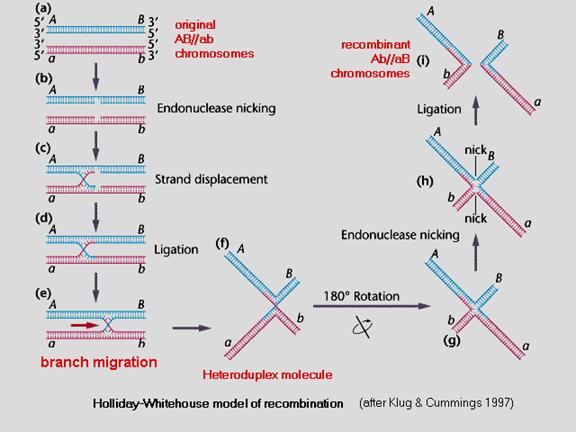
Genetic variation II
One more extension of Mendelian genetics
Morgan and Sturtevant – gene linkage
Goals
Morgan wanted to distinguish among the evolutionary theories of the Darwinists, neo-Lamarckists, and Mendelists such as de Vries
Methods – Drosophila genetics
Morgan began working with fruit flies (Drosophila melanogaster) in 1908 because he wanted a system for genetic analysis in which he could grow large numbers of individuals in small quarters quickly
Crosses
Crossed white-eyed mutant male with red-eyed female
F1progeny all red-eyed
What hypotheses can you make about inheritance of eye-color?
How many genes? Probably 1
How many alleles? Can’t tell
What allelic effects? Red allele dominant to white
Crossed the F1 sisters and brothers
F2 progeny
3 red-eyed:1white-eyed
Seems normal –
What hypotheses can we make about inheritance of eye color?
How many genes? Probably 1
How many alleles? 2 in this cross
What allelic effects? Red dominant to white
But, noticed something weird
All white-eyed flies were male!
What were ratios of sex and eye color?
Red-eyed flies were 50:50 male but white flies were 100% male
What hypotheses can we make about inheritance of eye color?
What would product law predict if sex and eye color were independently inherited?
Make a
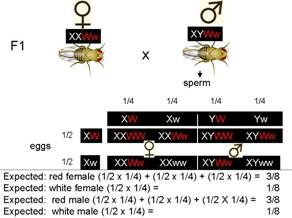
Observed:
½ red female:0 white female: ¼ red male:¼ white male
Sex and eye color do not follow product law
Thus, hypothesize that they are not inherited independently
Not independently segregating
Crossed F2 progeny and obtained white-eyed females
Bred them with white-eyed males to produce pure-breeding white-eyed line
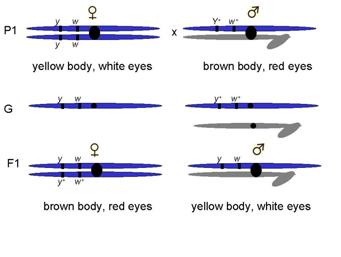
Reciprocal cross: crossed purebreeding white-eyed female with purebreeding red-eyed male
F1 progeny observed
½ red-eyed females : ½ white-eyed males Weird!!!
Recessive trait expressed in males
Crossed F1 siblings with each other
Outcome
¼ red-eyed female: ¼ white-eyed female: ¼ red-eyed male: ¼ white-eyed male
Explanation
Sex inherited by modified Mendelian mechanism
Females are XX (homogametic)
Males are XY (heterogametic)
Eye color
Eye color inherited by single gene, two allele system, red dominant to white
But, sex and eye color not inherited independently
Eye color is linked to sex
Eye color gene on X chromosome
So,
Males have only one eye color allele
Haploid
Females have two eye color alleles
Diploid
Ellen: What about dosage???
Called sex linkage
Gene or genes for sex determination are on same chromosome (X) as gene for eye color
Check out sex-linked inheritance problem set on web:
http://www.biology.arizona.edu/mendelian_genetics/problem_sets/sex_linked_inheritance_2/sex_linked_inheritance_2.html
More linkage
As Morgan and colleagues accumulated more mutants in their fly colony
They quickly discovered that linkage is not unique to X-chromosome
Found many linked traits on autosomes
Autosomes are all chromosomes that are not sex chromosomes
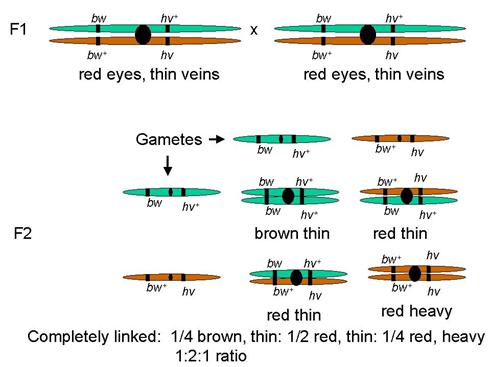
Example
Brown/Red eyes, Thin/Heavy veins
Two traits with genes both linked on same autosome
Predict: assort together
Phenotype frequencies: 1:2:1
Tightly linked
Tightly linked genes act like a single gene
In this case, like a single gene with codominant alleles
But, also found odd ratios
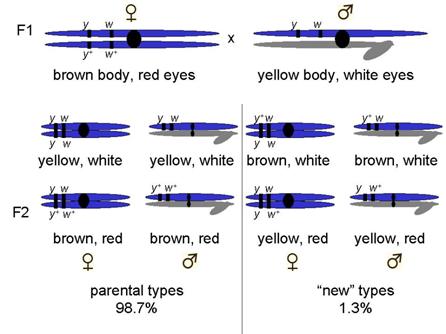
Example, yellow body and white eyes
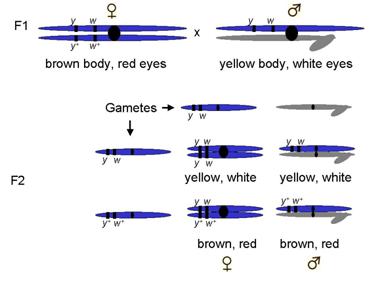
Crossed yellow-bodied, white-eyed males x brown-bodied, red-eyed females
F1 progeny as expected for sex-linked trait
What phenotypes do you expect?
Crossed F1s to get F2s
What ratios might be expected?
9:3:3:1 if unlinked
But, they knew they were both sex-linked and therefore on same chromosome
Linked to one another
So, expected what outcome?
1:1:1:1 ratio
¼ female yellow, white : ¼ female brown,red : ¼ male yellow, white : ¼ male brown,red
Most progeny fit that description
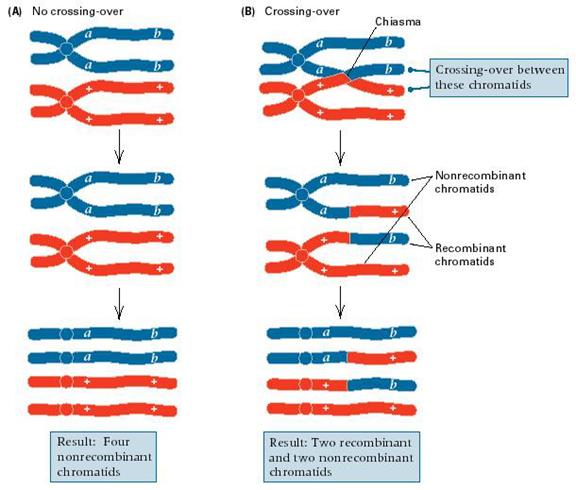
But a few did not!
Non-parental types