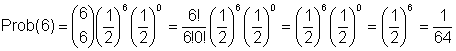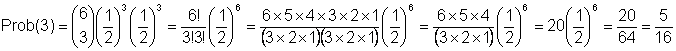Modeling drift over one generation of bottleneck
Our hypothetical example (from tussock moths after lupines die)
Parental population
With
Frequency of F alleles = p = 0.5
Frequency of S alleles = q = 0.5
Release gametes into gamete pool
Gamete frequencies
Same as parentals (no selection)
Frequency of F alleles = p = 0.5
Frequency of S alleles = q = 0.5
Assume that only N = 3 zygotes survive
Generate these 3 zygotes by sampling 2N gametes from the gamete pool
First, think of drift as a process of sampling gametes
What happens to allele frequencies?
In a diploid organism
Sampling gametes from gamete pool is a binomial process
Because there are only two possible outcomes to each trial
Either gamete will have F allele or it will have S allele
Binomial sampling process modeled by binomial probability
Remember binomial theorem from your homework assignment?
Binomial theorem states
Where
n = number of events = 2N gametes sampled
s = number of times a gamete has an F allele
(n-s) = t = number of times a gamete does not have an F allele
(and instead has an S allele)
For example,
What is probability that population will fix for F allele after this bottleneck down to a population of 3 caterpillars?
= Probability that all 3 zygotes will be homozygous FF
= Probability that all (2 x 3 = 6 gametes) will have F alleles
So,
n = number of events = 2N gametes sampled = 6
s = number of times a gamete has an F allele = 6
(n-s) = t = number of times a gamete does not have an F allele = 0
(and instead has an S allele)
A second example,
What is probability of obtaining one of each genotype?
= Probability that the three zygotes are (FF, FS, SS)
= Probability that 3 gametes have F allele and 3 gametes have S allele
So,
n = number of events = 2N gametes sampled = 6
s = number of times a gamete has an F allele = 3
(n-s) = t = number of times a gamete does not have an F allele = 3
(and instead has an S allele)
Now, think of drift as a process of sampling zygotes
What happens to allele frequencies?
In a diploid organism
Sampling zygotes from zygote pool is a trinomial process
Because there are three possible outcomes to each trial
Either a zygote will have FF genotype or it will have FS genotype or SS genotype
Trinomial sampling process modeled by trinomial probability
Trinomial theorem states
Where
n = number of events = N zygotes sampled
s = number of times a zygote has an FF genotype
t = number of times a zygote has an FS genotype
n-(s+t) = u = number of times a zygote does not have an FF or FS genotype
(and instead has an SS genotype)
Assume HW equilibrium
P = frequency of FF genotype
P = p2
H = frequency of FS genotype
H = 2pq
Q = frequency of SS genotype
Q = q2
For example,
What is probability that population will fix for F allele after this bottleneck down to a population of 3 caterpillars?
= Probability that all 3 zygotes will be homozygous FF
So,
n = number of events = N zygotes sampled = 3
s = number of times a zygote has an FF genotype= 3
t = number of times a zygote has an FS genotype = 0
n - (s+t) = u = number of times a zygote does not have either an FF or an FS genotype = 0
(and instead has an SS genotype)
Again, assume HW equilibrium
P = frequency of FF genotype
P = p2
H = frequency of FS genotype
H = 2pq
Q = frequency of SS genotype
Q = q2
A second example,
What is probability of obtaining one of each genotype?
= Probability that the three zygotes are (FF, FS, SS)
So,
n = number of events = N zygotes sampled = 3
s = number of times a zygote has an FF genotype= 1
t = number of times a zygote has an FS genotype = 1
n - (s+t) = u = number of times a zygote does not have either an FF or an FS genotype = 1
(and instead has an SS genotype)
Again, assume HW equilibrium
P = frequency of FF genotype
P = p2
H = frequency of FS genotype
H = 2pq
Q = frequency of SS genotype
Q = q2
Why is this result different from the one obtained by sampling gametes?
Because can get 3 F alleles and 3 S alleles in more ways than simply FF FS SS
Can also be from FS FS FS
A final example,
What is probability that our sample if 3 caterpillars will all be heterozygotes?
= Probability that all 3 zygotes will be homozygous FS
So,
n = number of events = N zygotes sampled = 3
s = number of times a zygote has an FF genotype= 0
t = number of times a zygote has an FS genotype = 3
n - (s+t) = u = number of times a zygote does not have either an FF or an FS genotype = 0
(and instead has an SS genotype)
Again, assume HW equilibrium
P = frequency of FF genotype
P = p2
H = frequency of FS genotype
H = 2pq
Q = frequency of SS genotype
Q = q2